I was surprised to discover that nearly three-quarters of our storage was near-zero float32s — and motivated to take advantage of this sparsity.
Background
At Sturdy Statistics, we build sparsity-inducing hierarchical mixture models for unsupervised and semi-supervised text organization. These models are extensions of topic models, with the ability to add arbitrary levels of hierarchy or additional branches, and to embed metadata into the structure of the graph. Our models learn a set of high level “topics” from the data, and then annotate the original data with these high level topics. We use DuckDB to store topic annotations at the paragraph and document level.
Because we explicitly model the power-law behavior of text data, our priors adopt a rich-get-richer scheme. Consequently, most topic activations are near zero. That means our data is inherently sparse — and ripe for storage optimization. There are a number of analytical benefits to this sparsity, but for the purpose of this post, the primary benefit is some sweet storage and speed wins.
Once I saw how sparse our data really was, I wanted to see how much space we could actually save in practice.
Sparsity
The next steps were
- Write arrays into DuckDB in a sparse format
- Write a set of wrappers that enable me to operate on these sparse arrays as if they are standard DuckDB lists
- Benchmark performance and storage wins (I’m optimistic)
For (1), I decided to use SciPy style sparse arrays (Dimension: int, indices: list(int8), Values: list(float32)). For example: [7, 0, 0, 0, 23] => (5, [1, 5], [7, 23])
Example Conversion in Numpy
import numpy as np
def numpy_to_sparse(x):
x = np.where(x < np.max(x)/3e3, 0.0, x).astype(np.uint8)
inds = np.where(x!=0)[0]
vals = x[inds]
dim = len(x)
vals = (dim, inds.astype(np.uint16), vals.astype(np.float32))
return vals
def numpy_from_sparse(vals):
dim = vals[0]
inds = vals[1]
data = vals[2]
arr = np.zeros(dim, dtype=np.float32)
arr[inds] = data
return arrWhen building wrapper functions, I had to choose between storing the sparse format in one column or splitting it into three. The benefit of storing it in one column is that I could fully abstract away the fact that these arrays are sparse. However, because DuckDB is a columnar data store, I opted to split out the indices and the values so I could flexibly access either component without reading both. Our data is sparse enough that the indices alone provide a mechanism to rapidly filter excerpts that contain a topic to some degree. If I stored the indices in the same column as the values, I would have to read 5× as much data (int8 vs int8+float32). The immediate performance benefit made the choice easy. I also fully removed the dimensionality parameter, opting to store it in the table metadata.
If my goal were to integrate this into DuckDB, I would have opted for the more ergonomic approach.
I put together a few helper functions that enabled me to perform all the operations. We use these functions internally, and we also publicly expose them in our API.
Sparse SQL Functions
CREATE OR REPLACE TEMPORARY MACRO sparse_list_extract(position, inds, vals) AS
COALESCE(vals[list_position(inds, position)], 0);
CREATE OR REPLACE TEMPORARY MACRO sparse_list_select(positions, inds, vals) AS
list_transform(positions, x-> sparse_list_extract(x, inds, vals));
CREATE OR REPLACE TEMPORARY MACRO dense_x_sparse_dot_product(arr, inds, vals) AS
list_dot_product(list_select(arr, inds)::DOUBLE[], vals::DOUBLE[]);
CREATE OR REPLACE TEMPORARY MACRO sparse_to_dense(K, inds, vals) AS
list_transform(
generate_series(1, K),
x-> coalesce(list_extract(vals, list_position(inds, x)), 0)
);These helpers replace array indexing, multi-indexing and converting sparse lists to dense. I was actually surprised how rarely I actually needed to perform this conversion: instead, for operations such as Hellinger distance or dot products, I can often restrict computation to the overlap in sparse indices instead (spoiler: performance wins are upcoming).
Benchmarking the Code
My first attempt was to benchmark DuckDB file sizes storing various levels of sparsity. I wanted a clear, granular understanding of how sparsity affects data at various levels of dimensionality and sparsity. I decided to put together some artificial data to benchmark the results…
Hint: this was a bad idea
I used numpy to generate a matrix of K×D where K is the number of topics (embedding dimension in Neural terms) and D is the number of documents. I set the first S rows to be 1/K and the rest were zero. I took these matrices and created two DuckDB files: one with raw storage and the other sparsely encoded.
I was shocked to see that there was virtually no difference in the file size. I was stunned that a 512-dimensional array occupied nearly the same space in DuckDB as two 8-dimensional arrays.
In my eagerness to get an easy 75% reduction in file size, I completely overlooked the fact that DuckDB applies Snappy compression to its files. Compressing long runs of zeros and ones is trivial for a compressor, so my synthetic data didn’t reveal much. Even though the synthetic benchmarks didn’t deliver the expected 75% reduction, they exposed how compression interacts with sparsity — a key insight for real-world data. I re-ran the test with random values in random positions and saw a modest but real improvement: ~20% average reduction.
Note: at this point in time my entire DuckDB file was a single database table consisting of a single list column, stored either raw or sparsely across two columns.
This was a solid win, albeit one much less than I hoped for. But it was enough to see this project through to the next stage.
Real Data (or the only benchmark that mattered)
I downloaded the DuckDB files from our publicly hosted indices to migrate to our sparse format for storage and performance benchmarking. I repurposed my previous script to read the original DuckDB files, convert the relevant list fields to a sparse format, and write a new file. The result made me simultaneously excited, confused — and hesitant:
This is a major reduction in our file sizes, hitting an over 50% reduction for some of our largest files.
What happened?
My initial test only measured a single column at a time. In production, however, we store several sparsely-populated arrays alongside any arbitrary metadata provided by our users or by our integrations. Since this metadata is not sparse, I had expected its presence would reduce the impact of our total file size reduction. But it had the opposite effect: we see a much more pronounced difference on real data. It appears that the additional columns handicapped the Snappy compression, rendering our sparse storage much more effective at the file level, not just the array level.
While we didn’t see the 75% reduction I had initially hoped for, compression still left us with a 52% average reduction across all files. I’ll take it!
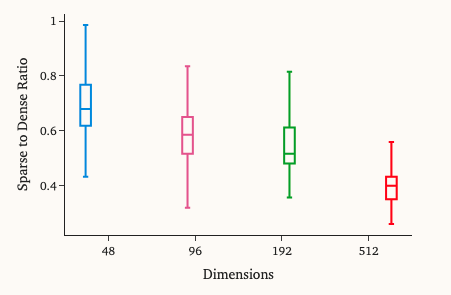
Scaling with Dimensionality
As expected, higher-dimensional arrays yielded larger storage reductions. Since our biggest datasets also have the highest topic counts, they benefited most.
While the large dataset performance is a big win, we care most about general performance because we support so many use cases:
- Our public Deep Dive Search creates a few hundred indices per day, split between our fast mode (48 dimensions) and our comprehensive mode (96 dimensions).
- Our production platform integrations have a slightly lower number of analyses trained per day with a default of 196 dimensions.
- Our large-scale models tend to be a more deliberate effort, with maybe 1–2 of these trained per week.
At the moment, the total amount of storage is relatively evenly split between these three categories of models. (And we hope these numbers go up as we begin to announce Sturdy Statistics publicly!)
Performance
I was both optimistic and concerned about the impact of sparse storage on performance. It turned out that we saw no observable changes in our search query performance or our topic retrieval performance. This was likely because I was able to leverage my dense_x_sparse_dot_product SQL function to avoid casting any sparse arrays to dense.
Our TopicSearch and TopicDiff however, initially saw significant performance degradations — about 2–5× slower. These APIs are much more analytical; they perform statistics over topic counts and expect dense arrays. I simply converted the sparse arrays to dense in my first pass, which obviously added overhead.
However, our new sparse storage format opened the door to fully leveraging UNNEST much more efficiently due to the lower dimensionality of the arrays and the separate indices-and-values arrays. Thanks to sparse storage, an UNNEST that previously expanded rows by 500× now expands them by just 3–8×.
This not only sped up DuckDB queries but also let me migrate a major portion of my Python post-processing directly into DuckDB. With some tinkering I was able to see a solid 2–10× performance improvement, with the larger improvements occurring on the larger datasets.
Sparse storage not only saved space—it made some operations up to 10× faster.
Conclusion
Benchmarks are just simulations; your own data tells the truth.
My main conclusion — one I’ve learned before and will likely learn again — is that the only benchmark that matters is the one on your own data. Simulations and estimates are helpful thought exercises; real workloads decide.
Second, while I initially tried to make sparse arrays feel “just like” dense arrays, embracing the different data structure is what unlocked most of the gains. UNNEST became powerful because indices and values live in separate columns. The “right” architecture wasn’t what I first expected; it emerged from benchmarking real use cases and looking carefully at what we were actually storing.
These sparse array functions live in a small GitHub repo, with documentation with examples. We plan to reach out to the DuckDB team soon to explore official support for sparse-style arrays. If you’re interested in running with this idea — for their own project or via open — I’d love to chat or help out however I can.